SPOTLIGHT: Yesterday, today and tomorrow @ Genomics and Bioinformatics Unit
Pubblicato il: 03.07.2019 18:10
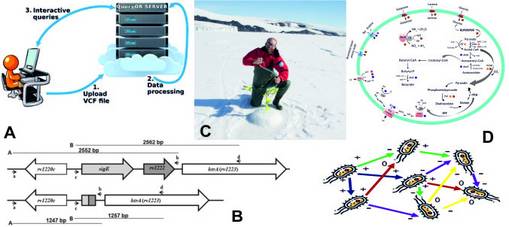
The Genomics and Bioinformatics Research Unit headed by Prof. Giorgio Valle deals with several aspects of genomic research, ranging from bioinformatics to metagenomics and large scale genomic analysis. In the last ten years, it specialized in Next Generation Sequencing (NGS)data analysis, including variant calling- annotation and prioritization, transcriptome, miRNA analysis and ChipSeq. Some of the latest developed tools are QueryOr (http://queryor.cribi.unipd.it/cgi-bin/queryor/mainpage.pl) and SCUBA (http://genomics.cribi.unipd.it/main/) that were designed for human variants analysis from exome and genome sequencing (Figure 1A). The bioinformatics group is also working on false-positive variants detection, a well-known issue in exome and genome analyses. This problem is due to the inclusion of rare alleles and sequencing errors in the human reference genome. Plant and Agro-Food Genomics is another topic covered by the Unit, with the aim of creating an integrated network of data, instruments and knowledge serving as a frame of reference for the scientific community.
Molecular microbiologyand Metagenomicsare two workhorses of the Unit, with several projects dealing with microbial genomics, anaerobic digestion, food-related and marine microbiology. Specific bioinformatics pipelines have been developed for microbial communities studies, such as the development of new methods for genome-centric metagenomics.
Molecular microbiology analyses are mainly focused on the study of sigma factors and G-quadruplexes in Mycobacterium tuberculosis, a research field investigated by Prof. Roberta Provvedi. Nucleic acids secondary structures may occur in single-stranded G-rich sequences and can epigenetically regulate cellular functions. Transcriptional profile investigations of M. tuberculosistreated with the G4 ligand BRACO-19 (G-quadruplexes stabilizer) are ongoing using RNAseq. This study includes also the regulatory network of SigE, one of the 13 sigma factors encoded by M. tuberculosis(Figure 1B). This sigma factor plays a very important role in virulence. For this reason, several mutants devoid of proteins required for SigE induction and activation were obtained in order to characterize the dynamics leading to SigE activation upon stress induction.
The study of the Venice lagoon is a very important research topic in marine microbiology. This work started as part of the Ocean Sampling Day (OSD) project, and it is still ongoing by means of collecting and analyzing water and sediment samples at different time points and different sites with the objective of characterizing the lagoon microbial community. Metagenetics, metagenomics and metatranscriptomics approaches were the main tools utilized. Moreover, two National Antarctic Research Program (PNRA) projects are investigating the microbial communities present in the Ross Sea (XXXIII Italian Antarctic Expedition), in collaboration with Istituto Nazionale di Oceanografia e di Geofisica Sperimentale (OGS), CNR Naples and University of Parma. The aims of these two studies are to understand prokaryotes interactions with Antarctic phytodetritus (Figure 1C) and to identify cold adapted enzymes for PUFA biosynthesis, respectively.
Another topic of the Unit is the investigation of Anaerobic Digestion (AD) microbiome(https://biogasmicrobiome.env.dtu.dk/ microbial-genomes.org/projects/biogasmicrobiome), which is under active development in collaboration with Prof. Irini Agelidaki of the DTU environment (https://www.dtu.dk/english/service/phonebook/person?id=4803&tab=1) and with Panagiotis Kougias of ELGO Demeter (Greece). Many projects related to this research topic are ongoing, aiming to a detailed characterization of the microbial community in methane-producing bioreactors by means of 16S rRNA sequencing and genome-centric metagenomics. One crucial aspect of this investigation is the creation of a global database including most of the genomes of the species involved; this process that has been recently boosted through an extensive application of the genome-centric approach (https://www.biorxiv.org/content/10.1101/680553v1). Another innovative study related to the AD system is focused on integration of biomass and wind power resulting in upgrading biogas to natural gas quality (i.e. methane) via hydrogen assisted anaerobic digestion. Additionally, genome-centric metatranscriptomics has been widely used to clarify the expressed genes and metabolic pathways. These tools are useful to decipher the complex microbial interactions driving the processes under investigation and as a basis to implement the Metabolic Flux Balance (MFB) analysis. In fact, a new MFB pipeline is now under development in order to study complex microbial communities (Figure 1D). The aim is to inspect the interactions occurring in a microbial community and clarify the effect of the environmental disturbances on the network of microbes.
People in the Genomics and Bioinformatics Unit: Prof. Giorgio Valle, Prof. Roberta Provvedi, Prof. Stefano Campanaro, Dr. Alessandro Vezzi, Dr.ssa Laura Treu, Dr. Davide Campagna, Dr. Marco Falda, Dr. Fabio De Pascale, Dr. Riccardo Schiavon, Dr.ssa Erika Feltrin, Dr.ssa Michela D'Angelo, Dr.ssa Rosanna Zimbello, Dr.ssa Arianna Basile.
If you like to know more about Genomics and Bioinformatics, visit us at:
www.instagram.com/bioinf.omics/





